经典遗传算法的Python实现
本文最后更新于 2025年6月4日 晚上
经典遗传算法的Python实现
理论
遗传算法(Genetic Algorithm, GA)由Prof.Holland 提出,是一种模拟生物染色体遗传行为结合达尔文生物进化理论的进化算法。它可以在给定的有限的搜索空间中找到全局最优解。
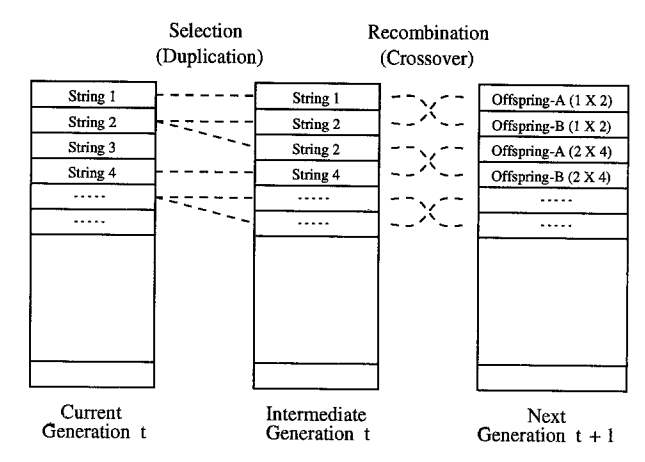
其运行过程如下图所示:
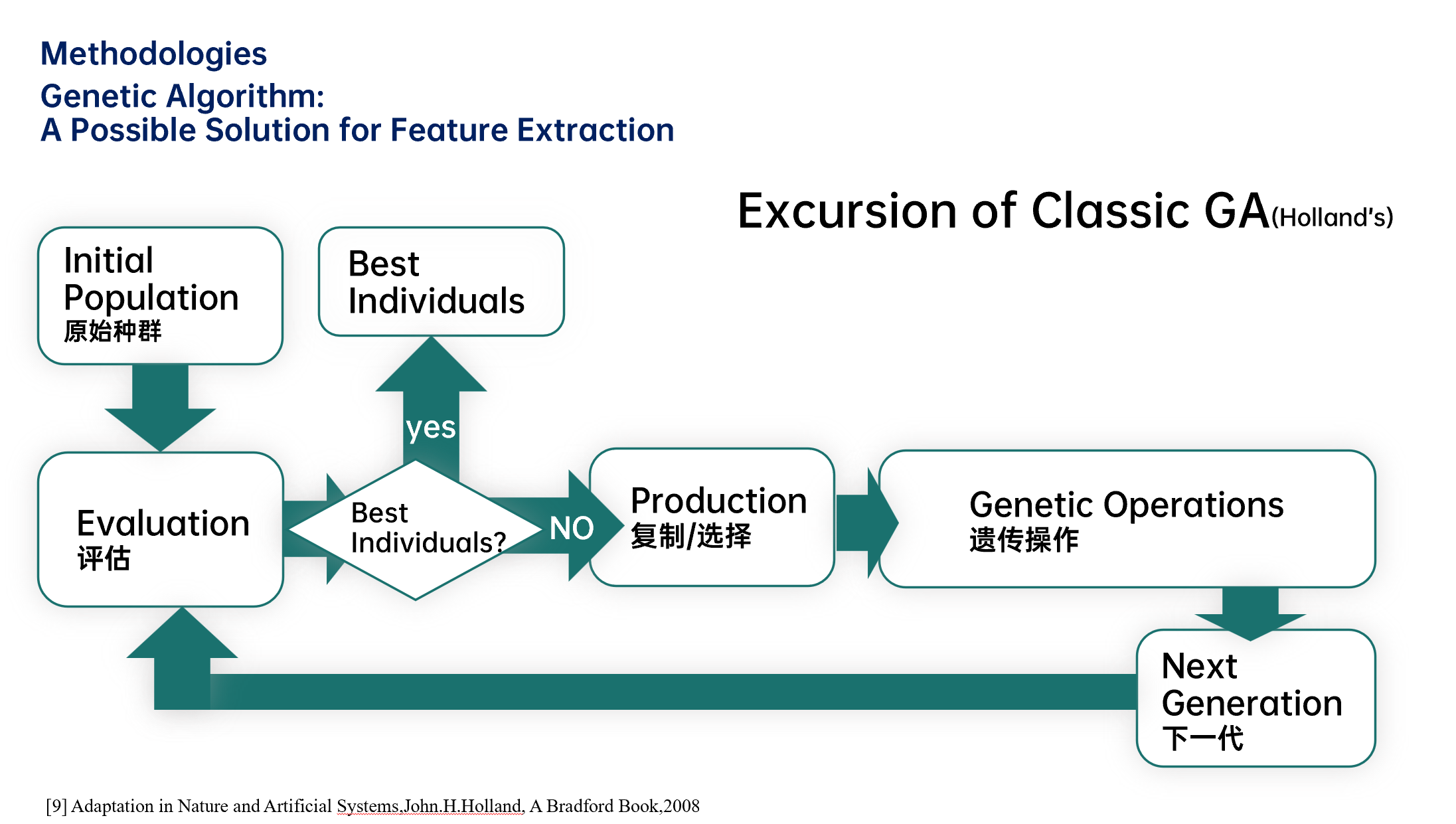
其运行的主要过程分为:种群初始化(population initialization)、评估(evaluation)、复制/选择(production)、遗传操作(genetic operation)四个模块,在Holland提出的经典遗传算法中遗传操作包括单点交叉(single point crossover)和突变(mutation)两步。
概念
遗传算法中一个问题的可能解是目标函数包括的所有自变量的对应的一个特定值的集合,称为个体(individual)。在遗传算法中,这些值被线性排列,称为一条染色体(chromosome)。一个个体对应一条染色体,所有个体的染色体长度应当是相等的。染色体上描述某个变量对应值的区域被称为该变量在这个染色体上对应的基因(gene)。
遗传算法中,个体的集合被称为种群(population),所有可能个体的集合是遗传算法的搜索空间(searching space)。
运行遗传算法前的准备:搜索空间的确定
搜索空间主要是通过为目标问题的所有可能解决方案确定一种通用的编码方式来实现。在经典遗传算法中,这一编码机制为二进制字符串,称为位串(string)。每一个个体都通过某种方式转化为唯一的一个位串。
这种转化的方式要求:
- 搜索空间内的个体数量是有限的。
- 编码后所有个体在搜索空间中都是均匀分布的。
- 对位串中对每个基因的一次改变的对搜索进度的影响程度是相同的。
种群初始化
由于实际问题中遗传算法要面临的状态编码数量非常庞大,算法需要给定一些个体以便从它们开始查找最优解。通常,这些个体通过随机抽样的方式被确定,随机抽样得到的个体集合称为初始种群(initial population)。
评估
评估是量化个体优劣性的方式。所谓个体的优劣性即个体对应的目标问题的一个解对目标问题解决程度的描述。评估是通过将编码后的个体进行解码,然后送入一个可以量化个体优劣性的函数中实现的。这个函数被称为适应度函数(fitness function)。每个个体经过适应度函数后会得到一个表征其性能的值,称为适应度(fitness),\(f\)。
种群中个体\(i\)的正规化适应度(normalized fitness)定义为:
\[\frac{f_i}{\overline{f}}\]
其中,\(f_i\)表示评估函数对第\(i\)个个体的评估结果,\(\overline{f}\)表示种群的平均评估。
个体的适应度越高,表明这个状态对应的系统结果越能够符合我们的要求。
复制·选择(随机余数采样)
选择后的每个个体的适应度格式为x.xx,即有小数部分和整数部分。适应度的整数部分表示该个体会被复制多少次。 复制中的选择机制:
这样就能直接地让优秀的个体获得更多被复制的机会,而适应度整数部分为0的个体因为不会被复制而被淘汰。
但是,那些适应度比较低的个体中仍然可能有对系统有益的部分,为了尽可能地保留这些部分,遗传算法在选择阶段还规定:
对所有的个体,适应度的小数部分表示额外被复制的概率。
如此,每一个个体中对系统有益的部分都能够被尽可能地复制。
比如,适应度2.3的个体能够获得2次复制,并且有0.3的概率能获得第三次复制的机会。
这样的机制能够用数学表示为:
\[M(H,t+1)=M(H,t)\frac{f(H,t)}{\overline{f}}\]
其中M表示的是种群中的一个亚种(Sub-populations)。
总结:
选择的过程即为有概率地对种群中的个体进行复制,可以发现,适应度越高的个体被复制的概率就越大。
原始种群经过复制后形成中间种(Intermediate Generation)。
重组(单点交叉)
遗传算法中重组的本质是杂交(Crossover),其过程主要有两步:
- 随机地使得个体间两两配对。
- 随机地选取一对个体,两者在某个随机且相同的比特位处断开,前后的两段基因型进行交叉互换。
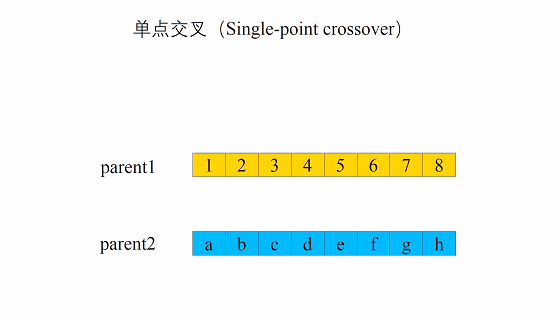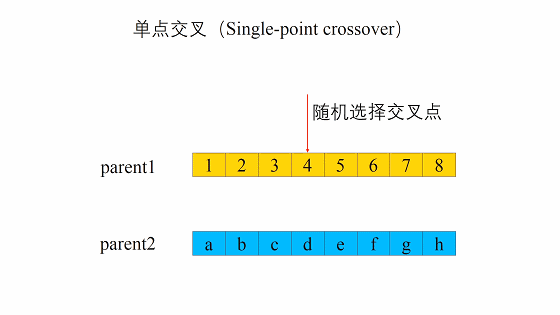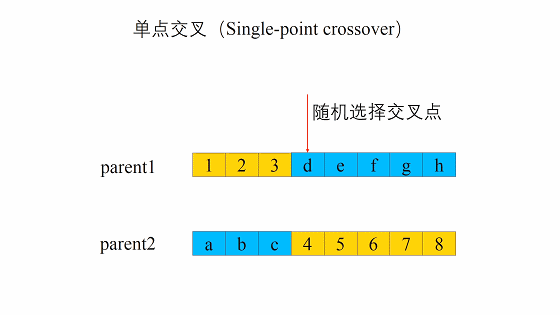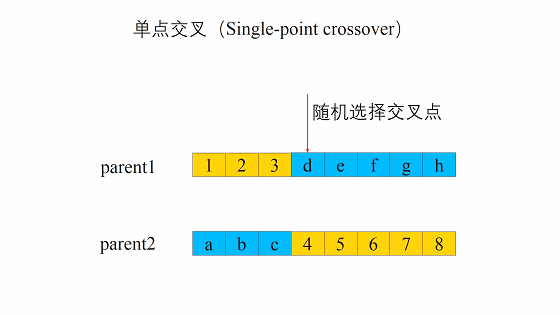
新生成的两个个体称为后代(Offspring),后代能够插入到下一代的概率计作\(p_c\)。
突变(反转突变)
重组之后利用突变算子对后代作突变处理,对于种群中的所有比特位,其有\(p_m\)的概率发生比特反转。同遗传学一样,突变概率一个非常小的概率,通常小于1%。中间种经过重组和突变,最终能称为新的种群。
Python 代码实现
在Python中,设计每一个只含有0和1的列表(list)表示一个个体,一个种群对应一个二维列表。
初始种群生成
初始种群生成的函数能够完成的功能是:对于给定的初始种群数量和染色体长度,可以随机地生成一些个体。
基本思路是利用嵌套的for循环实现这一功能:内层的for循环用于为每一个个体列表随机填满0和1(random.randint(0, 1)),外层的for循环用于将生成好的个体放入种群中。
1 | |
转义和评估
转义
要想评估每一个个体的优劣性,需要按照模板将位串对应区域的比特转换为十进制数,如下图所示,这一步称为转义(translation)。
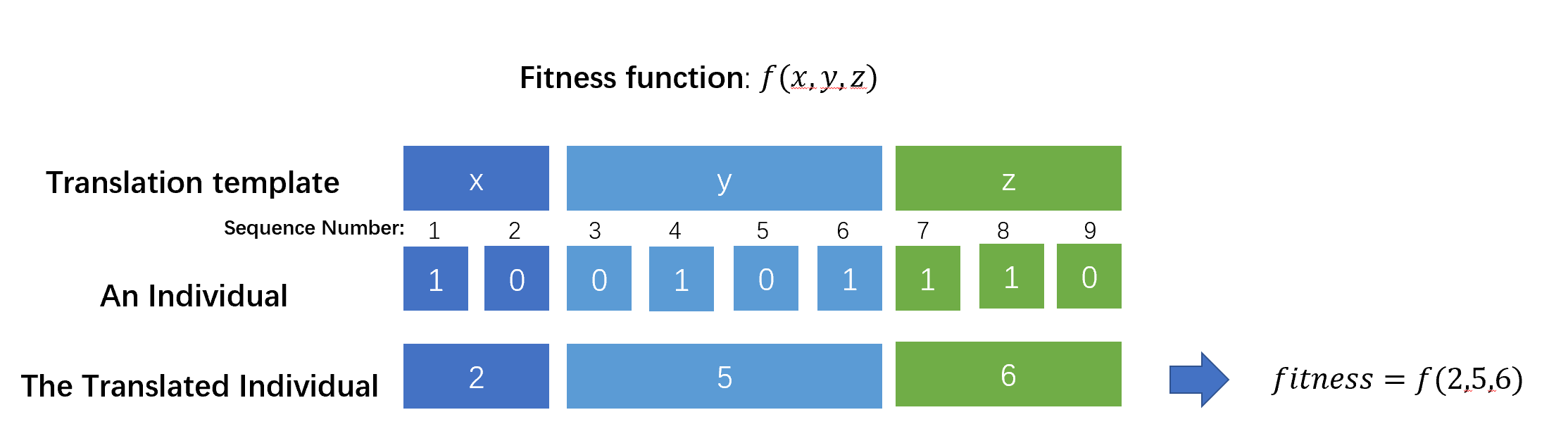
转义的基因模板可以通过给定一个数组gene_pattern,其中标有每个基因的截断位置实现。例如:gene_pattern = [3,6]表示indiv[1]到indiv[3]视为一个基因;indiv[4]到indiv[6]视为另一个基因。
使用for循环和列表切片将位串按照gene_pattern给定的位置进行划分,并手动去除掉[],’,’和空格。
1
2
3
4
5
6# 根据gene_pattern划分基因
for k in range(len(gene_pattern)):
string.append((''.join(
str(population[i][gene_pattern[k - 1]:gene_pattern[k]]))
).replace('[', '').replace(']', '').replace(
',', '').replace(' ', ''))gene_pattern[k - 1]:gene_pattern[k]对k=0不适用,因此还需要手动将第一个基因替换为0:gene_pattern[0]的切片。
1
2
3
4# 第一个基因转义后为'[]',需要手动替换为0:gene_pattern[0]的切片
string[0] = ''.join(str(population[i][0:gene_pattern[0]])
.replace('[', '').replace(
']', '').replace(',', '').replace(' ', ''))
最后使用int函数将每个基因转化为对应的十进制。
1
gene_trans = int(string[m], 2)
强烈建议基因的长度应当相等,如此目标问题可能解在整个搜索空间中的分布才是均匀的,这样才满足模式定理(schema theory)的基本成立条件。
1 | |
评估
将转义后的个体带入到适应度函数中就可以得到该个体所对应的适应度。 其中,适应度函数使用fitness_func进行定义。
传入该函数的是一个列表,函数中各变量的值通过访问列表中对应的元素获取。下面给出了一个适应度函数\(f(x_0,x_1,x_2,x_3)=x_0+x_1+(2x_2+3x_3)^2\)的例子:
1
2
3
4# fitness_function.py
def fitness_func(x):
y= x[0] + x[1]+ pow((2*(x[2]))+(3*(x[3])),2)
return float(y)fitness,该列表的序号和种群中population个体对应的序号相同,因此可以根据序号查询到每个个体对应的适应度。
获得适应度后,需要对每个个体适应度除以当前种群适应度的均值以正规化每个个体的适应度:indiv_fitness_norm = fitness[j] / avg_fitness.
其中avg_fitness = sum(fitness) / float(len(fitness))是当前种群的适应度的平均值,该值也会被该函数返回以便在主函数中观察进化过程。
在实现中,加入了其他几个调试参数,比如用于是否输出正规化适应度的fitness_normalization,以及是否打印调试输出的debug_print。
1 | |
选择
根据经典遗传算法提到的理论,每个个体正规化后的适应度的整数部分代表个体出现在下一轮的种群中的次数。
1
2
3for j in range(int(fitness_norm[i])):
population_inter.append(population[i])
copy_times = copy_times + 1
概率以随机生成一个\([0,1]\)的随机数与其小数部分比较来表示。
1 | |
1 | |
单点交叉
在进行单点交叉之前,首先需要检查当前种群中的个体数是否大于1,如果当前种群里面只有1个个体,那么交叉将不会进行,并且返回一个布尔值False以便让主程序终止进化过程。
1
2
3
4
5
6if population_size == 1: # 检查当前种群中是否只有一个个体
# 输出告警
print('【Warning】only one indiv in current population')
return population, False
else:
print('single crossover crossover rate:', crossover_rate)
然后对当前种群中的所有个体两两配对,这个概念通过shuffle打乱所有个体的序号和在for循环处设置range(0, population_size - 1, 2)以每两个一组实现。
对于每一组个体,使用cross_point = random.randint(0,chromesome_length - 1)随机选择一个交叉点。
然后进行单点交叉,其基本思路是创建两个列表parent1和parent2,对于每个列表,分别放入其中一个个体交叉点前的位串和另一个个体交叉点后的位串。
1 | |
1 | |
突变
突变的基本思路与交叉基本相同:遍历每个个体,并用random.random() < mutation_rate模拟突变概率。对于每个个体,随机选择一个突变点的位置:
1
mutation_point = random.randint(0, chromesome_length - 1)
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15def bit_inverse(b: 'int'):
'''
Function:
inverse the bit.
Parameters:
b - int
'''
# if this element is 1, then change to be 0.
if b == 1:
b = 0
else:
# if this element is 0, then change to be 1.
b = 1
return b
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32def mutation(population: 'list', mutation_rate: '0<= float<= 1') -> 'list':
'''
Function:
do the mutation based on the mutation rate.
Parameters:
population - two dimension list
mutation_rate - 0<=float<=1, the rate of crossover.
Return:
population - two dimension list
'''
population_size = len(population)
chromesome_length = len(population[0])
print('mutation mutation rate:', mutation_rate)
for i in range(population_size):
# chech each indiv with mutation rate
if random.random() < mutation_rate:
# random select the mutation point
mutation_point = random.randint(0, chromesome_length - 1)
print('indiv:', i, ' chromosome:', population[i],
' mutated: yes', ' mutation point:', mutation_point)
population[i][mutation_point] = bit_inverse(population[i][mutation_point])
print('---after mutation---')
print('indiv:', i, ' chromosome:', population[i],
' mutated: yes')
else:
print('indiv:', i, ' chromosome:', population[i],
' mutated: not')
print('========')
return population
示例程序
调用写好的GA库:
1
2import GeneticAlgorithm as GA
import matplotlib.pyplot as plt
1
m = GA.initia_population(10, 10)avg_fitness为0则停止迭代,因此avg_fitness初始化为1.
1
2
3
4avg_fitness = 1
epoches = 0
fitness = []
flag = True
设置迭代,根据算法的运行流程每个迭代中需要完成:
- 将当前种群中的所有个体进行转义,得到种群
m_t - 评估每个个体,并且得到对应的适应度列表
m_f以及当前种群的平均适应度avg_fitness - 根据适应度列表
m_f对种群m进行选择 - 对选择后的个体进行单点交叉,单点交叉的发生概率为0.9
- 对选择后的个体进行突变,突变的发生概率为0.1
1 | |
整个程序的终止条件有2个:
- 算法无法找到适应度大于1200的个体,在这种情况下设置算法运行1000轮迭代后自行停止,
epoches == 1000。
- 在某次选择后种群只剩下一个个体,无法完成交叉,利用
single_crossover函数中的判断条件,设置flag == False时终止运行。
1 | |
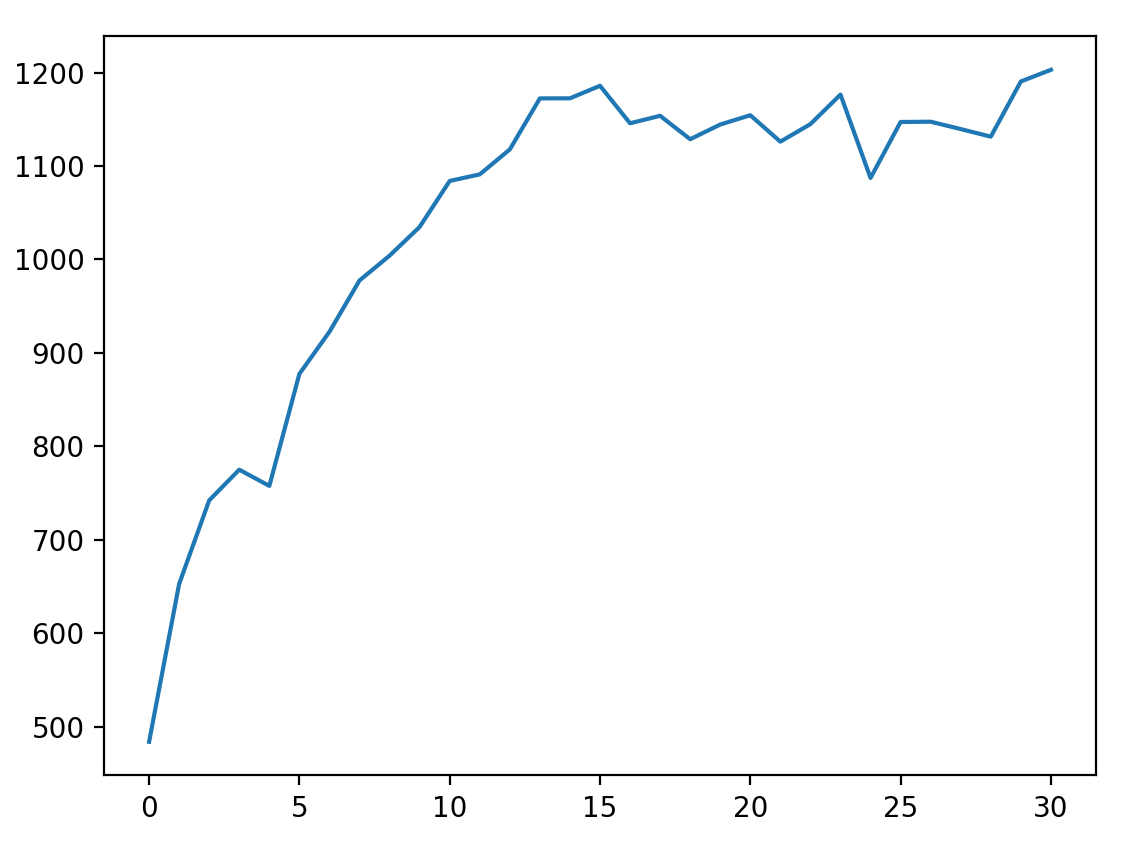
算法结束后打印epoches等变量以及最后一代的全部个体。并绘制整个运行过程中的avg_fitness与epoches的变化图,以便检查程序的运行情况。
1
2
3
4
5
6
7
8
9print('finish')
print('=============================')
print('epoches: ', epoches)
print('max fitness: ', max(fitness))
print('winners:')
for i in range(len(m)):
print('indiv: ',i,' chromosome:',m[i])
plt.plot(fitness)
plt.show()
运行结果
程序每一轮的输出如下所示:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155
156
157
158
159
160
161
162
163
164
165
166
167
168
169
170
171
172
173
174
175
176
177
178
179
180
181
182
183
184
185
186
187
188
189
190
191
192
193
194
195
196
197
198
199
200
201
202
203
204
205
206
207
208
209initializing the population
indiv:* 0 chromesome: [0, 1, 1, 1, 1, 0, 0, 0, 0, 1]
indiv:* 1 chromesome: [0, 0, 0, 1, 1, 1, 0, 0, 1, 1]
indiv:* 2 chromesome: [0, 0, 1, 1, 0, 0, 1, 1, 0, 0]
indiv:* 3 chromesome: [1, 1, 1, 0, 1, 1, 1, 1, 0, 0]
indiv:* 4 chromesome: [1, 0, 1, 0, 1, 0, 0, 0, 1, 1]
indiv:* 5 chromesome: [1, 1, 0, 1, 1, 0, 0, 0, 0, 1]
indiv:* 6 chromesome: [0, 0, 0, 0, 1, 1, 0, 1, 0, 1]
indiv:* 7 chromesome: [0, 1, 0, 0, 0, 1, 1, 1, 1, 1]
indiv:* 8 chromesome: [1, 1, 1, 1, 1, 1, 1, 1, 0, 0]
indiv:* 9 chromesome: [1, 0, 1, 0, 1, 1, 1, 0, 1, 0]
indiv:* 10 chromesome: [1, 1, 1, 0, 0, 1, 1, 0, 0, 0]
indiv:* 11 chromesome: [0, 0, 1, 1, 1, 1, 1, 1, 0, 1]
indiv:* 12 chromesome: [1, 1, 1, 1, 1, 1, 1, 1, 0, 0]
indiv:* 13 chromesome: [0, 1, 0, 0, 0, 1, 1, 0, 1, 0]
indiv:* 14 chromesome: [1, 1, 0, 1, 1, 1, 1, 1, 1, 0]
indiv:* 15 chromesome: [1, 1, 1, 0, 1, 1, 1, 1, 0, 1]
indiv:* 16 chromesome: [0, 1, 1, 1, 0, 0, 1, 1, 1, 1]
indiv:* 17 chromesome: [1, 0, 0, 0, 1, 1, 0, 1, 0, 0]
indiv:* 18 chromesome: [0, 1, 0, 0, 1, 1, 0, 0, 1, 1]
indiv:* 19 chromesome: [0, 0, 1, 1, 0, 0, 1, 0, 1, 1]
======Round: 0 ==============
translation
indiv: 0 chromosome: [1, 3, 4, 1]
indiv: 1 chromosome: [0, 1, 6, 3]
indiv: 2 chromosome: [0, 3, 1, 4]
indiv: 3 chromosome: [3, 2, 7, 4]
indiv: 4 chromosome: [2, 2, 4, 3]
indiv: 5 chromosome: [3, 1, 4, 1]
indiv: 6 chromosome: [0, 0, 6, 5]
indiv: 7 chromosome: [1, 0, 3, 7]
indiv: 8 chromosome: [3, 3, 7, 4]
indiv: 9 chromosome: [2, 2, 7, 2]
indiv: 10 chromosome: [3, 2, 3, 0]
indiv: 11 chromosome: [0, 3, 7, 5]
indiv: 12 chromosome: [3, 3, 7, 4]
indiv: 13 chromosome: [1, 0, 3, 2]
indiv: 14 chromosome: [3, 1, 7, 6]
indiv: 15 chromosome: [3, 2, 7, 5]
indiv: 16 chromosome: [1, 3, 1, 7]
indiv: 17 chromosome: [2, 0, 6, 4]
indiv: 18 chromosome: [1, 0, 6, 3]
indiv: 19 chromosome: [0, 3, 1, 3]
evaluation
successfully load fitness function
indiv: 0 fitness: 125.0
indiv: 1 fitness: 442.0
indiv: 2 fitness: 199.0
indiv: 3 fitness: 681.0
indiv: 4 fitness: 293.0
indiv: 5 fitness: 125.0
indiv: 6 fitness: 729.0
indiv: 7 fitness: 730.0
indiv: 8 fitness: 682.0
indiv: 9 fitness: 404.0
indiv: 10 fitness: 41.0
indiv: 11 fitness: 844.0
indiv: 12 fitness: 682.0
indiv: 13 fitness: 145.0
indiv: 14 fitness: 1028.0
indiv: 15 fitness: 846.0
indiv: 16 fitness: 533.0
indiv: 17 fitness: 578.0
indiv: 18 fitness: 442.0
indiv: 19 fitness: 124.0
average fitness: 483.65
indiv: 0 normal fitness: 0.25845135945415076
indiv: 1 normal fitness: 0.913884007029877
indiv: 2 normal fitness: 0.41145456425100796
indiv: 3 normal fitness: 1.4080430063062133
indiv: 4 normal fitness: 0.6058099865605293
indiv: 5 normal fitness: 0.25845135945415076
indiv: 6 normal fitness: 1.5072883283366072
indiv: 7 normal fitness: 1.5093559392122404
indiv: 8 normal fitness: 1.4101106171818465
indiv: 9 normal fitness: 0.8353147937558152
indiv: 10 normal fitness: 0.08477204590096145
indiv: 11 normal fitness: 1.7450635790344258
indiv: 12 normal fitness: 1.4101106171818465
indiv: 13 normal fitness: 0.29980357696681487
indiv: 14 normal fitness: 2.1255039801509357
indiv: 15 normal fitness: 1.7491988007856922
indiv: 16 normal fitness: 1.1020365967124988
indiv: 17 normal fitness: 1.195079086115993
indiv: 18 normal fitness: 0.913884007029877
indiv: 19 normal fitness: 0.25638374857851753
selection
indiv:* 0 normal fitness: 0.25845135945415076 No extra copy
indiv:* 1 normal fitness: 0.913884007029877 Copy times: 1 extra
indiv:* 2 normal fitness: 0.41145456425100796 Copy times: 1 extra
indiv:* 3 normal fitness: 1.4080430063062133 Copy times: 1
indiv:* 3 normal fitness: 1.4080430063062133 Copy times: 2 extra
indiv:* 4 normal fitness: 0.6058099865605293 No extra copy
indiv:* 5 normal fitness: 0.25845135945415076 No extra copy
indiv:* 6 normal fitness: 1.5072883283366072 Copy times: 1
indiv:* 6 normal fitness: 1.5072883283366072 Copy times: 2 extra
indiv:* 7 normal fitness: 1.5093559392122404 Copy times: 1
indiv:* 7 normal fitness: 1.5093559392122404 No extra copy
indiv:* 8 normal fitness: 1.4101106171818465 Copy times: 1
indiv:* 8 normal fitness: 1.4101106171818465 No extra copy
indiv:* 9 normal fitness: 0.8353147937558152 Copy times: 1 extra
indiv:* 10 normal fitness: 0.08477204590096145 No extra copy
indiv:* 11 normal fitness: 1.7450635790344258 Copy times: 1
indiv:* 11 normal fitness: 1.7450635790344258 Copy times: 2 extra
indiv:* 12 normal fitness: 1.4101106171818465 Copy times: 1
indiv:* 12 normal fitness: 1.4101106171818465 Copy times: 2 extra
indiv:* 13 normal fitness: 0.29980357696681487 No extra copy
indiv:* 14 normal fitness: 2.1255039801509357 Copy times: 1
indiv:* 14 normal fitness: 2.1255039801509357 Copy times: 2
indiv:* 14 normal fitness: 2.1255039801509357 No extra copy
indiv:* 15 normal fitness: 1.7491988007856922 Copy times: 1
indiv:* 15 normal fitness: 1.7491988007856922 No extra copy
indiv:* 16 normal fitness: 1.1020365967124988 Copy times: 1
indiv:* 16 normal fitness: 1.1020365967124988 No extra copy
indiv:* 17 normal fitness: 1.195079086115993 Copy times: 1
indiv:* 17 normal fitness: 1.195079086115993 No extra copy
indiv:* 18 normal fitness: 0.913884007029877 Copy times: 1 extra
indiv:* 19 normal fitness: 0.25638374857851753 Copy times: 1 extra
single crossover crossover rate: 0.9
shuffle
========
indiv: 0 chromosome: [1, 0, 1, 0, 1, 1, 1, 0, 1, 0] crossovered: not
indiv: 1 chromosome: [0, 0, 1, 1, 1, 1, 1, 1, 0, 1] crossovered: not
========
indiv: 2 chromosome: [1, 1, 0, 1, 1, 1, 1, 1, 1, 0] crossovered: yes
indiv: 3 chromosome: [0, 0, 0, 1, 1, 1, 0, 0, 1, 1] crossovered: yes
---after crossover---
indiv: 2 chromosome: [1, 1, 0, 1, 1, 1, 1, 0, 1, 1] crossovered: yes
indiv: 3 chromosome: [0, 0, 0, 1, 1, 1, 0, 1, 1, 0] crossovered: yes
========
indiv: 4 chromosome: [1, 1, 1, 0, 1, 1, 1, 1, 0, 0] crossovered: yes
indiv: 5 chromosome: [0, 0, 0, 0, 1, 1, 0, 1, 0, 1] crossovered: yes
---after crossover---
indiv: 4 chromosome: [1, 0, 0, 0, 1, 1, 0, 1, 0, 1] crossovered: yes
indiv: 5 chromosome: [0, 1, 1, 0, 1, 1, 1, 1, 0, 0] crossovered: yes
========
indiv: 6 chromosome: [0, 1, 0, 0, 1, 1, 0, 0, 1, 1] crossovered: not
indiv: 7 chromosome: [0, 0, 1, 1, 1, 1, 1, 1, 0, 1] crossovered: not
========
indiv: 8 chromosome: [1, 1, 0, 1, 1, 1, 1, 1, 1, 0] crossovered: not
indiv: 9 chromosome: [1, 1, 1, 1, 1, 1, 1, 1, 0, 0] crossovered: not
========
indiv: 10 chromosome: [0, 1, 1, 1, 0, 0, 1, 1, 1, 1] crossovered: yes
indiv: 11 chromosome: [1, 1, 1, 1, 1, 1, 1, 1, 0, 0] crossovered: yes
---after crossover---
indiv: 10 chromosome: [0, 1, 1, 1, 0, 0, 1, 1, 1, 0] crossovered: yes
indiv: 11 chromosome: [1, 1, 1, 1, 1, 1, 1, 1, 0, 1] crossovered: yes
========
indiv: 12 chromosome: [0, 0, 1, 1, 0, 0, 1, 0, 1, 1] crossovered: not
indiv: 13 chromosome: [1, 1, 1, 1, 1, 1, 1, 1, 0, 0] crossovered: not
========
indiv: 14 chromosome: [0, 1, 0, 0, 0, 1, 1, 1, 1, 1] crossovered: yes
indiv: 15 chromosome: [1, 1, 1, 0, 1, 1, 1, 1, 0, 0] crossovered: yes
---after crossover---
indiv: 14 chromosome: [0, 1, 0, 0, 0, 1, 1, 1, 0, 0] crossovered: yes
indiv: 15 chromosome: [1, 1, 1, 0, 1, 1, 1, 1, 1, 1] crossovered: yes
========
indiv: 16 chromosome: [1, 1, 1, 0, 1, 1, 1, 1, 0, 1] crossovered: yes
indiv: 17 chromosome: [0, 0, 0, 0, 1, 1, 0, 1, 0, 1] crossovered: yes
---after crossover---
indiv: 16 chromosome: [1, 1, 1, 0, 1, 1, 0, 1, 0, 1] crossovered: yes
indiv: 17 chromosome: [0, 0, 0, 0, 1, 1, 1, 1, 0, 1] crossovered: yes
========
indiv: 18 chromosome: [0, 0, 1, 1, 0, 0, 1, 1, 0, 0] crossovered: yes
indiv: 19 chromosome: [1, 0, 0, 0, 1, 1, 0, 1, 0, 0] crossovered: yes
---after crossover---
indiv: 18 chromosome: [0, 0, 0, 0, 1, 1, 0, 1, 0, 0] crossovered: yes
indiv: 19 chromosome: [1, 0, 1, 1, 0, 0, 1, 1, 0, 0] crossovered: yes
mutation mutation rate: 0.1
indiv: 0 chromosome: [1, 0, 1, 0, 1, 1, 1, 0, 1, 0] mutated: not
========
indiv: 1 chromosome: [0, 0, 1, 1, 1, 1, 1, 1, 0, 1] mutated: not
========
indiv: 2 chromosome: [1, 1, 0, 1, 1, 1, 1, 0, 1, 1] mutated: not
========
indiv: 3 chromosome: [0, 0, 0, 1, 1, 1, 0, 1, 1, 0] mutated: not
========
indiv: 4 chromosome: [1, 0, 0, 0, 1, 1, 0, 1, 0, 1] mutated: not
========
indiv: 5 chromosome: [0, 1, 1, 0, 1, 1, 1, 1, 0, 0] mutated: not
========
indiv: 6 chromosome: [0, 1, 0, 0, 1, 1, 0, 0, 1, 1] mutated: not
========
indiv: 7 chromosome: [0, 0, 1, 1, 1, 1, 1, 1, 0, 1] mutated: not
========
indiv: 8 chromosome: [1, 1, 0, 1, 1, 1, 1, 1, 1, 0] mutated: not
========
indiv: 9 chromosome: [1, 1, 1, 1, 1, 1, 1, 1, 0, 0] mutated: not
========
indiv: 10 chromosome: [0, 1, 1, 1, 0, 0, 1, 1, 1, 0] mutated: not
========
indiv: 11 chromosome: [1, 1, 1, 1, 1, 1, 1, 1, 0, 1] mutated: not
========
indiv: 12 chromosome: [0, 0, 1, 1, 0, 0, 1, 0, 1, 1] mutated: not
========
indiv: 13 chromosome: [1, 1, 1, 1, 1, 1, 1, 1, 0, 0] mutated: not
========
indiv: 14 chromosome: [0, 1, 0, 0, 0, 1, 1, 1, 0, 0] mutated: not
========
indiv: 15 chromosome: [1, 1, 1, 0, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 16 chromosome: [1, 1, 1, 0, 1, 1, 0, 1, 0, 1] mutated: not
========
indiv: 17 chromosome: [0, 0, 0, 0, 1, 1, 1, 1, 0, 1] mutated: not
========
indiv: 18 chromosome: [0, 0, 0, 0, 1, 1, 0, 1, 0, 0] mutated: not
========
indiv: 19 chromosome: [1, 0, 1, 1, 0, 0, 1, 1, 0, 0] mutated: not
========
最后一轮的输出如下:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140
141
142
143
144
145
146
147
148
149
150
151
152
153
154
155======Round: 30 ==============
translation
indiv: 0 chromosome: [2, 1, 7, 7]
indiv: 1 chromosome: [3, 2, 7, 7]
indiv: 2 chromosome: [1, 2, 6, 7]
indiv: 3 chromosome: [3, 0, 7, 7]
indiv: 4 chromosome: [3, 2, 7, 7]
indiv: 5 chromosome: [3, 0, 7, 6]
indiv: 6 chromosome: [1, 3, 7, 7]
indiv: 7 chromosome: [3, 1, 7, 7]
indiv: 8 chromosome: [1, 3, 7, 7]
indiv: 9 chromosome: [3, 1, 7, 7]
indiv: 10 chromosome: [1, 3, 7, 7]
indiv: 11 chromosome: [1, 3, 7, 7]
indiv: 12 chromosome: [1, 3, 7, 7]
evaluation
successfully load fitness function
indiv: 0 fitness: 1228.0
indiv: 1 fitness: 1230.0
indiv: 2 fitness: 1092.0
indiv: 3 fitness: 1228.0
indiv: 4 fitness: 1230.0
indiv: 5 fitness: 1027.0
indiv: 6 fitness: 1229.0
indiv: 7 fitness: 1229.0
indiv: 8 fitness: 1229.0
indiv: 9 fitness: 1229.0
indiv: 10 fitness: 1229.0
indiv: 11 fitness: 1229.0
indiv: 12 fitness: 1229.0
average fitness: 1202.923076923077
indiv: 0 normal fitness: 1.0208466555825553
indiv: 1 normal fitness: 1.0225092722854585
indiv: 2 normal fitness: 0.9077887197851388
indiv: 3 normal fitness: 1.0208466555825553
indiv: 4 normal fitness: 1.0225092722854585
indiv: 5 normal fitness: 0.8537536769407853
indiv: 6 normal fitness: 1.021677963934007
indiv: 7 normal fitness: 1.021677963934007
indiv: 8 normal fitness: 1.021677963934007
indiv: 9 normal fitness: 1.021677963934007
indiv: 10 normal fitness: 1.021677963934007
indiv: 11 normal fitness: 1.021677963934007
indiv: 12 normal fitness: 1.021677963934007
selection
indiv:* 0 normal fitness: 1.0208466555825553 Copy times: 1
indiv:* 0 normal fitness: 1.0208466555825553 No extra copy
indiv:* 1 normal fitness: 1.0225092722854585 Copy times: 1
indiv:* 1 normal fitness: 1.0225092722854585 No extra copy
indiv:* 2 normal fitness: 0.9077887197851388 Copy times: 1 extra
indiv:* 3 normal fitness: 1.0208466555825553 Copy times: 1
indiv:* 3 normal fitness: 1.0208466555825553 No extra copy
indiv:* 4 normal fitness: 1.0225092722854585 Copy times: 1
indiv:* 4 normal fitness: 1.0225092722854585 No extra copy
indiv:* 5 normal fitness: 0.8537536769407853 Copy times: 1 extra
indiv:* 6 normal fitness: 1.021677963934007 Copy times: 1
indiv:* 6 normal fitness: 1.021677963934007 No extra copy
indiv:* 7 normal fitness: 1.021677963934007 Copy times: 1
indiv:* 7 normal fitness: 1.021677963934007 No extra copy
indiv:* 8 normal fitness: 1.021677963934007 Copy times: 1
indiv:* 8 normal fitness: 1.021677963934007 No extra copy
indiv:* 9 normal fitness: 1.021677963934007 Copy times: 1
indiv:* 9 normal fitness: 1.021677963934007 No extra copy
indiv:* 10 normal fitness: 1.021677963934007 Copy times: 1
indiv:* 10 normal fitness: 1.021677963934007 No extra copy
indiv:* 11 normal fitness: 1.021677963934007 Copy times: 1
indiv:* 11 normal fitness: 1.021677963934007 Copy times: 2 extra
indiv:* 12 normal fitness: 1.021677963934007 Copy times: 1
indiv:* 12 normal fitness: 1.021677963934007 No extra copy
single crossover crossover rate: 0.9
shuffle
========
indiv: 0 chromosome: [0, 1, 1, 0, 1, 1, 0, 1, 1, 1] crossovered: yes
indiv: 1 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] crossovered: yes
---after crossover---
indiv: 0 chromosome: [0, 1, 1, 0, 1, 1, 1, 1, 1, 1] crossovered: yes
indiv: 1 chromosome: [0, 1, 1, 1, 1, 1, 0, 1, 1, 1] crossovered: yes
========
indiv: 2 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] crossovered: yes
indiv: 3 chromosome: [1, 0, 0, 1, 1, 1, 1, 1, 1, 1] crossovered: yes
---after crossover---
indiv: 2 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] crossovered: yes
indiv: 3 chromosome: [1, 0, 0, 1, 1, 1, 1, 1, 1, 1] crossovered: yes
========
indiv: 4 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] crossovered: yes
indiv: 5 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] crossovered: yes
---after crossover---
indiv: 4 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] crossovered: yes
indiv: 5 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] crossovered: yes
========
indiv: 6 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] crossovered: not
indiv: 7 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] crossovered: not
========
indiv: 8 chromosome: [1, 1, 0, 0, 1, 1, 1, 1, 1, 1] crossovered: yes
indiv: 9 chromosome: [1, 1, 0, 0, 1, 1, 1, 1, 1, 0] crossovered: yes
---after crossover---
indiv: 8 chromosome: [1, 1, 0, 0, 1, 1, 1, 1, 1, 0] crossovered: yes
indiv: 9 chromosome: [1, 1, 0, 0, 1, 1, 1, 1, 1, 1] crossovered: yes
========
indiv: 10 chromosome: [1, 1, 0, 1, 1, 1, 1, 1, 1, 1] crossovered: yes
indiv: 11 chromosome: [1, 1, 1, 0, 1, 1, 1, 1, 1, 1] crossovered: yes
---after crossover---
indiv: 10 chromosome: [1, 1, 0, 0, 1, 1, 1, 1, 1, 1] crossovered: yes
indiv: 11 chromosome: [1, 1, 1, 1, 1, 1, 1, 1, 1, 1] crossovered: yes
========
indiv: 12 chromosome: [1, 1, 0, 1, 1, 1, 1, 1, 1, 1] crossovered: not
indiv: 13 chromosome: [1, 1, 1, 0, 1, 1, 1, 1, 1, 1] crossovered: not
mutation mutation rate: 0.1
indiv: 0 chromosome: [0, 1, 1, 0, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 1 chromosome: [0, 1, 1, 1, 1, 1, 0, 1, 1, 1] mutated: not
========
indiv: 2 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 3 chromosome: [1, 0, 0, 1, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 4 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 5 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 6 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 7 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 8 chromosome: [1, 1, 0, 0, 1, 1, 1, 1, 1, 0] mutated: not
========
indiv: 9 chromosome: [1, 1, 0, 0, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 10 chromosome: [1, 1, 0, 0, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 11 chromosome: [1, 1, 1, 1, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 12 chromosome: [1, 1, 0, 1, 1, 1, 1, 1, 1, 1] mutated: not
========
indiv: 13 chromosome: [1, 1, 1, 0, 1, 1, 1, 1, 1, 1] mutated: not
========
finish
=============================
epoches: 31
max fitness: 1202.923076923077
winners:
indiv: 0 chromosome: [0, 1, 1, 0, 1, 1, 1, 1, 1, 1]
indiv: 1 chromosome: [0, 1, 1, 1, 1, 1, 0, 1, 1, 1]
indiv: 2 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1]
indiv: 3 chromosome: [1, 0, 0, 1, 1, 1, 1, 1, 1, 1]
indiv: 4 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1]
indiv: 5 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1]
indiv: 6 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1]
indiv: 7 chromosome: [0, 1, 1, 1, 1, 1, 1, 1, 1, 1]
indiv: 8 chromosome: [1, 1, 0, 0, 1, 1, 1, 1, 1, 0]
indiv: 9 chromosome: [1, 1, 0, 0, 1, 1, 1, 1, 1, 1]
indiv: 10 chromosome: [1, 1, 0, 0, 1, 1, 1, 1, 1, 1]
indiv: 11 chromosome: [1, 1, 1, 1, 1, 1, 1, 1, 1, 1]
indiv: 12 chromosome: [1, 1, 0, 1, 1, 1, 1, 1, 1, 1]
indiv: 13 chromosome: [1, 1, 1, 0, 1, 1, 1, 1, 1, 1]
运行过程中的avg_fitness与epoches关系如下: